Background
AlphaFold is an AI system developed by Google DeepMind that predicts a protein’s 3D structure from its amino acid sequence. It regularly achieves accuracy competitive with experiment.
Google DeepMind and EMBL’s European Bioinformatics Institute (EMBL-EBI) have partnered to create AlphaFold DB to make these predictions freely available to the scientific community. The latest database release contains over 200 million entries, providing broad coverage of UniProt (the standard repository of protein sequences and annotations). We provide individual downloads for the human proteome and for the proteomes of 47 other key organisms important in research and global health. We also provide a download for the manually curated subset of UniProt (Swiss-Prot).
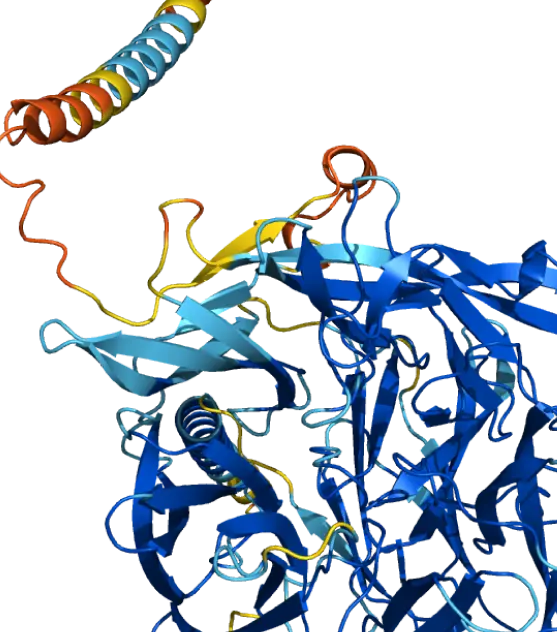
Q8I3H7: May protect the malaria parasite against attack by the immune system. Mean pLDDT 85.57.
In CASP14, AlphaFold was the top-ranked protein structure prediction method by a large margin, producing predictions with high accuracy. While the system still has some limitations, the CASP results suggest AlphaFold has immediate potential to help us understand the structure of proteins and advance biological research.
Let us know how the AlphaFold Protein Structure Database has been useful in your research, or if you have questions not answered in the FAQs, at alphafold@deepmind.com.
If your use case isn't covered by the database, you can generate your own AlphaFold predictions using this open source code, which also supports multimer prediction.
What’s new?
Community datasets - February 2026
The AlphaFold Protein Structure Database is now accepting large-scale datasets from scientific communities with deep domain expertise. This move aims to empower specialist communities to contribute and address remaining gaps in structural knowledge, particularly for organisms with limited sequence data.
The first such datasets already available include: 17M+ bacterial predictions from the AllTheBacteria project, parasite structures from the Wheeler Lab, and 435,000+ predictions from the Big Fantastic Virus Database (BFVD) and Viro3D.
What’s next?
We plan to continue updating the database with structures for newly discovered protein sequences, and to improve features and functionality in response to user feedback. Please follow Google DeepMind's and EMBL-EBI’s social channels for updates.
Licence and attributions
Data is available for academic and commercial use, under a CC-BY-4.0 licence.
EMBL-EBI expects attribution (e.g., in publications, services, or products) for any of its online services, databases, or software in accordance with good scientific practice.
If you use this resource, please cite the following papers:
Note: A given structure may be associated with further publications. For authoritative information regarding the relevant publication(s), please consult the dataset collection or the structure metadata provided on the entry page.
If you use data from AlphaMissense in your work, please cite the following paper:
AlphaFold Data Provided by GDM:
AlphaFold Data Copyright (2022) DeepMind Technologies Limited.
For AlphFold Data provided by third party data providers:
- Kinetoplastid data copyright (2025) Wheeler Lab
Wheeler RJ. A resource for improved predictions of Trypanosoma and Leishmania protein three-dimensional structure. PLoS One (2021) - AllTheBacteria data copyright (2025) AllTheBacteria Consortium
Hunt M, Lima L, Anderson D, Bouras G, Hall M, Hawkey J, Schwengers O, Shen W, Lees JA, Zamin Iqbal Z. BioRXiV (2025) - BVFD Data Copyright (2025) The BFVD Development Team
Kim, RS et al. BFVD—a large repository of predicted viral protein structures. NAR (2024) - Viro3D dataset (2025)
Litvin, U et al. Viro3D: a comprehensive database of virus protein structure predictions. Mol Syst Biol (2025)
AlphaMissense Copyright (2023) DeepMind Technologies Limited.
EMBL-EBI training
Recorded webinar
Accessing and interpreting predicted protein structures from AlphaFold database
Online tutorial